Projects
Here are projects I have developed, the links refer to the Github repositories where the code is freely available and the corresponding jupyter (.ipynb) notebook contains extensive markdown to explain the motivation, development and conclusions of the project. I enjoy working through projects from beginning to end, so in each of those project I start by conceptualising the question, collecting the data, cleaning, analysing, modelling, interpretation and giving future perspectives.
-
Analysis of tweets involving the hashtag “#UN”: Analysis of distribution and sentiment of user response through tweets pertaining to major political events.
Techniques: API data collection, NLP sentiment classification, Time series analysis, PCA, Hierarchical Clustering, Data visualisation.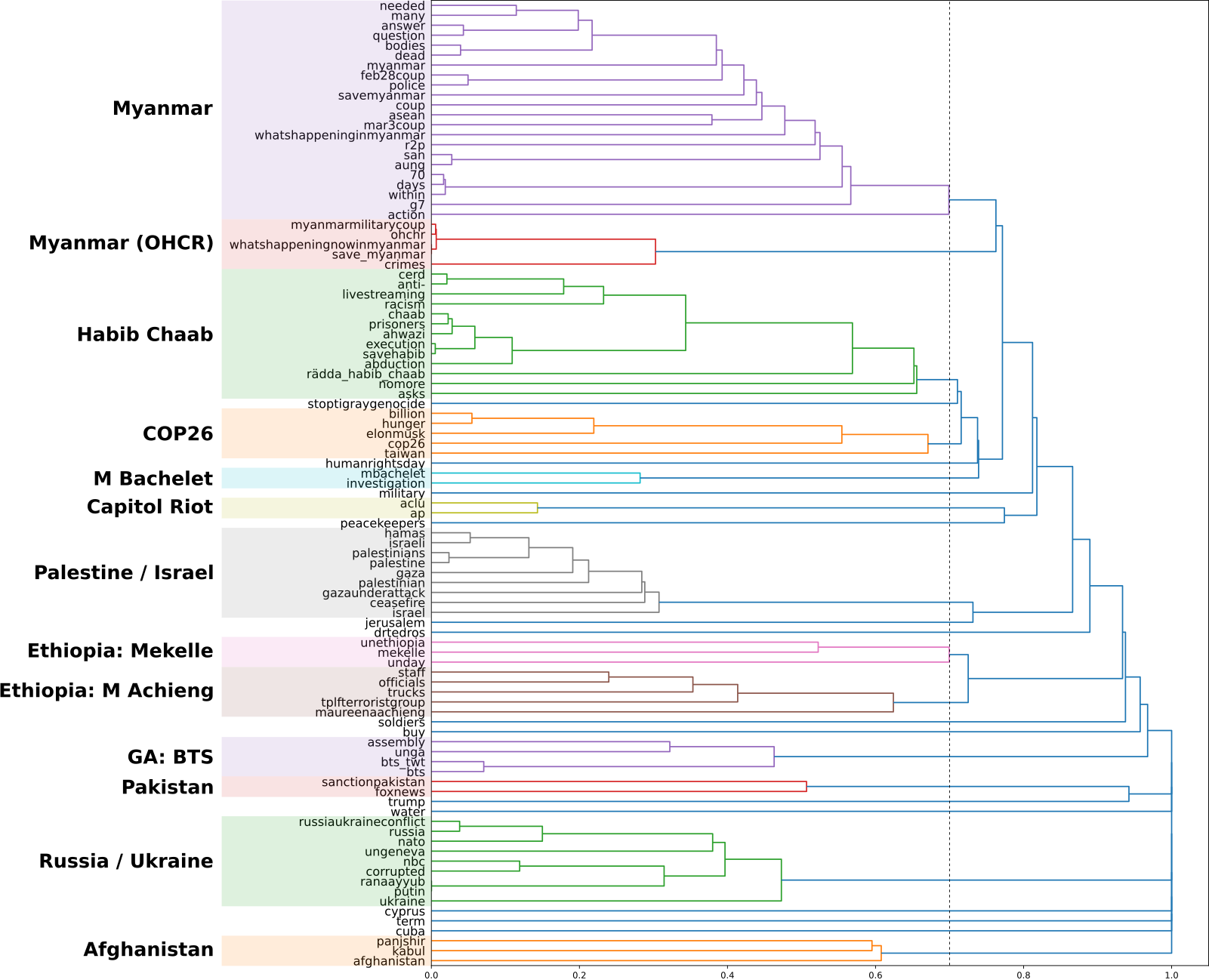
-
Calculating beer volume in a glass: Model trained using data from webcam using Convolutional Neural Networks to provide a regression to beer volume.
Techniques: Data collection, Convolutional Neural Networks.
Academic projects: These have been developed in an academic context using a combination of Mathematica, python, R, Matlab and Shell, as suitable for each purpose. See the explanations below and corresponding publications:
-
Compilation of all mathematical and physical advances in modelling pre-implantation embryonic development. We performed an in detail revision of the extensive literature of the field, went through the modelling approaches and synthesised the knowledge into a concise article. Topics covered are dynamical systems theory, agent based modelling, single-cell sequencing, stochastic differential equations and data visualisation.
See: Herrera-Delgado E and Maître JL†. (2021). A theoretical understanding of mammalian preimplantation development. Cells Dev. 203752. PDF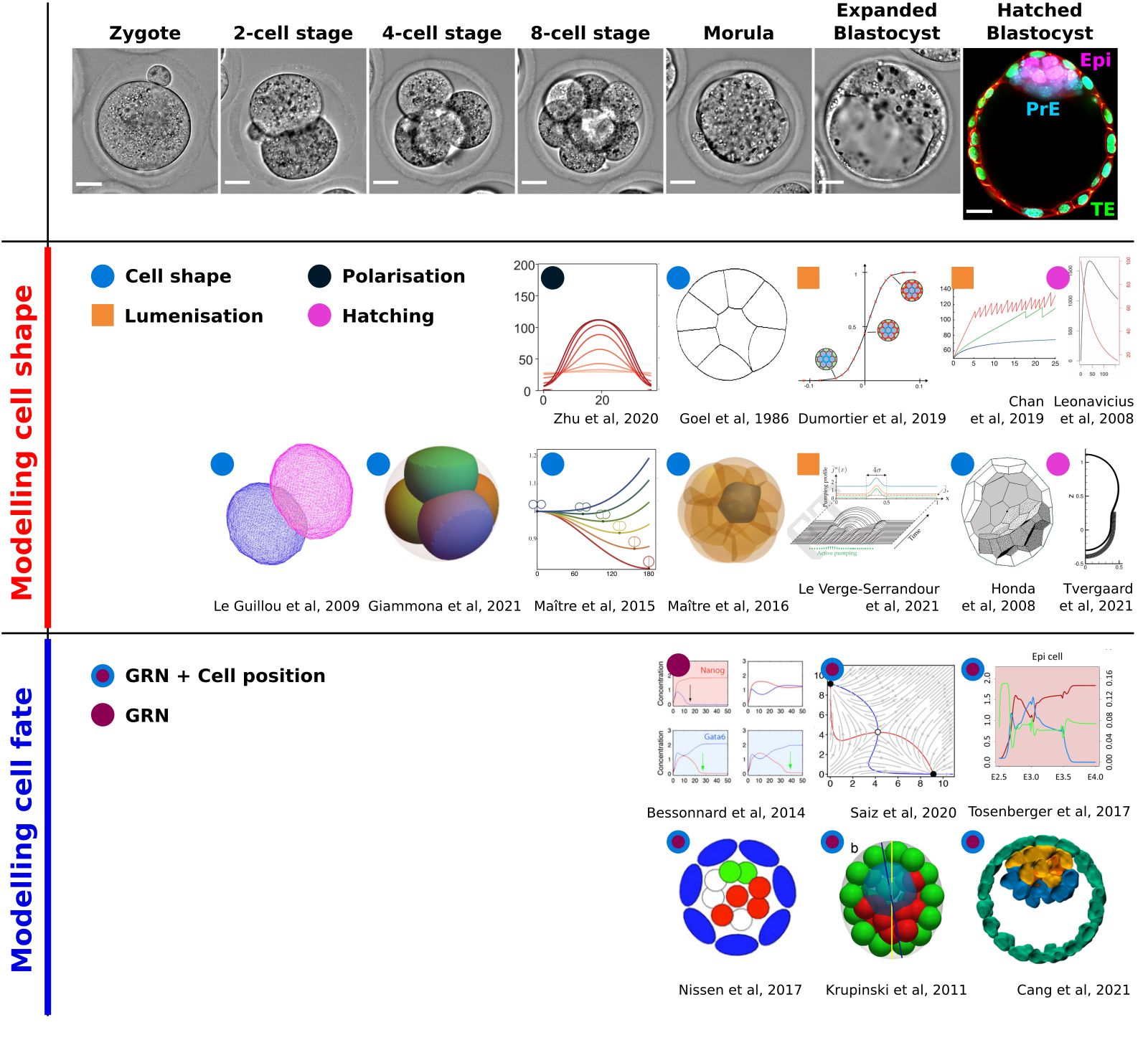
-
Project discovering for the first time how living tissues establish sharp boundaries during development, e.g. between different organs. Highly collaborative project over which I took leadership. Working with theoretical physicists and experimental biologists to solve this complex problem. Extensive iteration between myself and the experimental scientists to create a tailored and accurate model, while consulting with the theoreticians, the model was tested experimentally, iterating this process until finding the end solution. Mains tools: Gaussian Naive Bayes, Image Analysis, SDEs, Data Clustering, Dynamical Systems Theory, Data Visualisation, Storytelling.
See: Exelby K*, Herrera-Delgado E*†, Garcia Perez L, Perez-Carrasco R, Sagner A, Metzis V, Sollich P† and Briscoe J†. (2021). Precision of Tissue Patterning is Controlled by Dynamical Properties of Gene Regulatory Networks. Development. 10.1242/dev.197566. PDF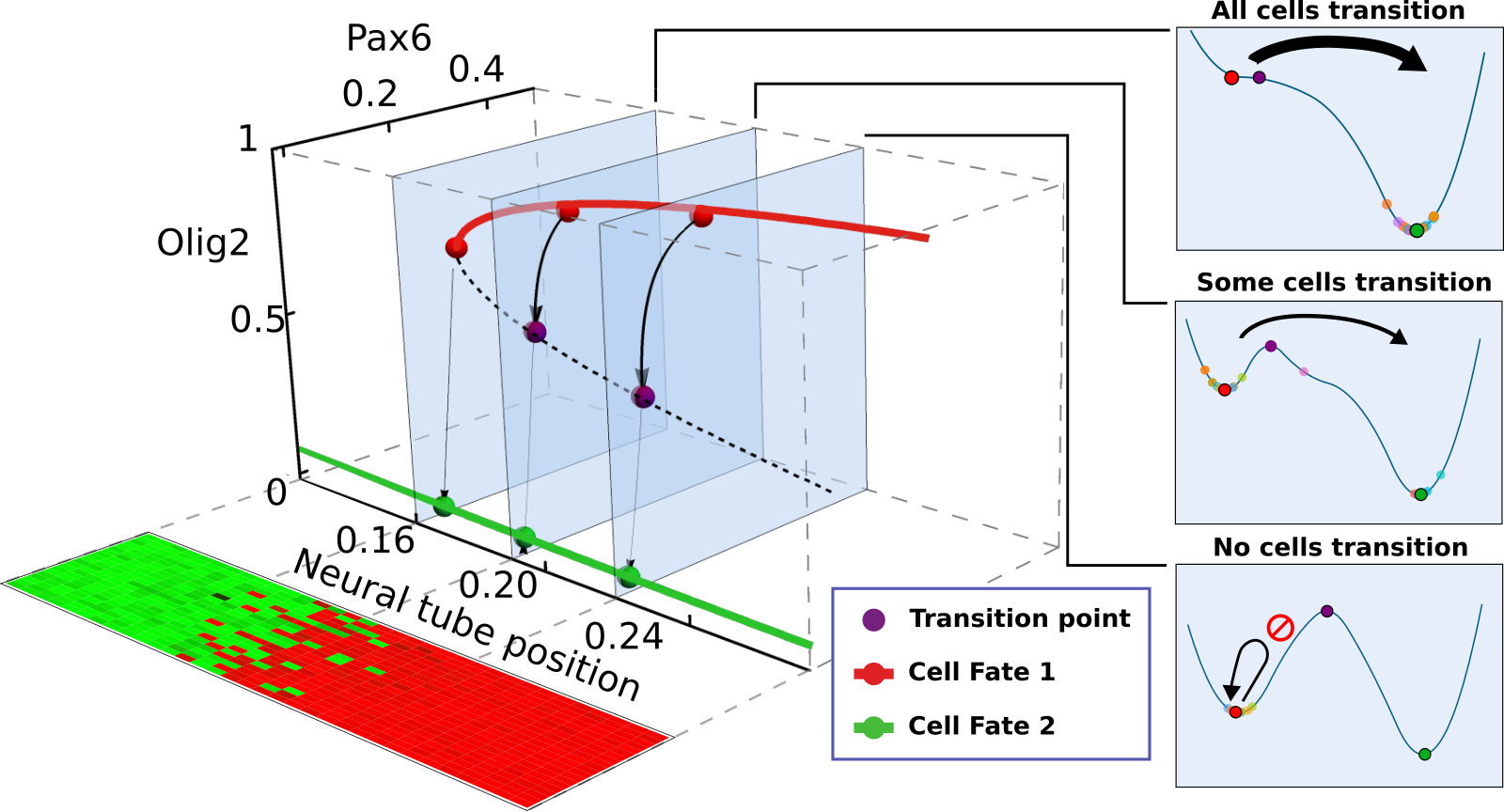
-
Mathematical technique for dimensionality reduction of dynamical models to aid with user interpretability. Project grounded on theoretical physics and applied mathematics, where we developed a method based on classical techniques to simplify models represented as networks. We used this technique to answer questions regarding how classical biological networks function that would otherwise not be possible to tackle. Main tools: Projection techniques, Matrix Calculus, Memory functions, Perturbation theory, Data visualisation, Storytelling.
See: Herrera-Delgado E, Briscoe J† and Sollich P†. (2020). Tractable nonlinear memory functions as a tool to capture and explain dynamical behaviors. Phys Rev Research. 10.1103/PhysRevResearch.2.043069. PDF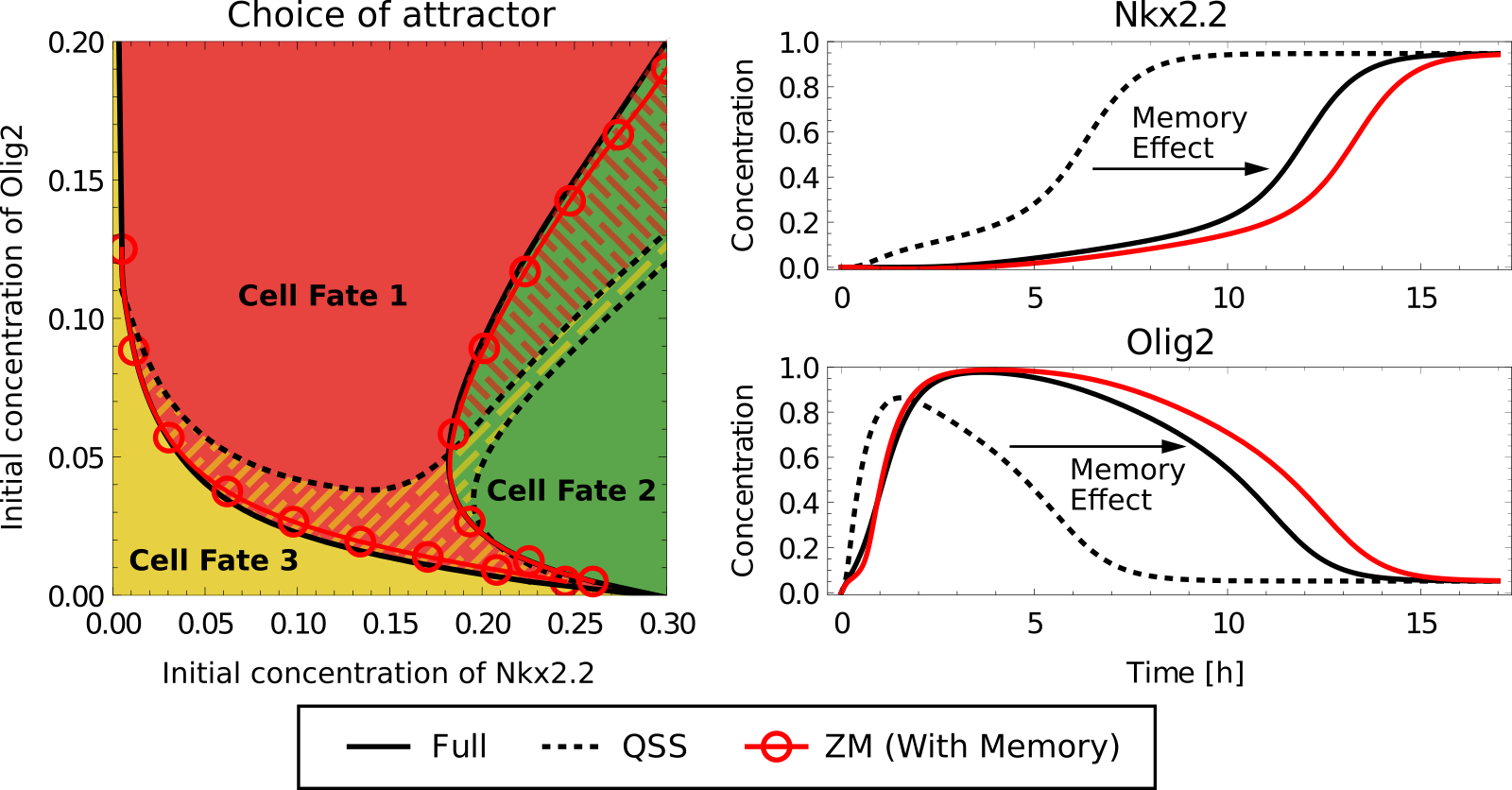
-
Deconstruction of memory functions to find the most important features of a model. We used classical mechanics to construct models that can be simplified into memory functions. We then analysed those memory functions to find key components of the networks. Main tools: Projection techniques, Matrix Calculus, Data visualisation, Storytelling.
See: Herrera-Delgado E, Perez-Carrasco R, Briscoe J† and Sollich P†. (2018). Memory Functions Reveal Structural Properties of Gene Regulatory Networks. PLoS Comput Biol. 14(2):e1006003. PDF